In this Chemistry Magazine,you have chemistry related news about a molecularly imprinted polymers (MIP) for sarafloxacin was prepared by the use of itaconic acid as the functional monomer and ethylene glycol dimethacrylate as the crosslinking monomer. But we will also show you the funny side of chemistry, as we earlier discussed the games we stumbled on. Why don’t you take a free test at the casino laboratory by discovering the best casino bonus chips at freecasinogameschips.com? You will find many games that will satisfy your scientific potential. Not let’s go back to the real topic, shall we? To improve selectivity, cobalt(II) ions was used to form metal-template complex, which provided the specific binding sites similar to metal-chelate antibody. The influences of different anions and cations on the recognition performance of the MIP were investigated in protic solvent. Due to the specific structure of the complex of cobalt(II)-sarafloxacin, it increased the specific selectivity of the polymer to sarafloxacin. In addition, we founded that anions also promoted the molecular recognition, leading to a further increased the specific selectivity of the polymer. Scatchard analysis show that two classes of binding sites existed in the metal complex imprinted polymers, with their maximum apparent binding capacity was estimated to be 51.3 mmol g-1 and 32.6 mmol g-1, respectively. Over the traditional organic phase synthesis of MIP, the present protocol was achieved in aqueous media, keeping the consistent of the preparation process and recognition process of the MIP. The results on substrate selectivity of imprinted polymer revealed that the prepared MIP had better binding affinity for template than the other tested compounds. Before we dive into the scientific research, we would like to redirect you to a review of one of the best gaming platform. While Lion Casino has reopened and is better than ever, with new features, games and very attractive offers.
Effect of cations on the recognition of sarafloxacin-co(II) complexes with imprinted polymer
A molecularly imprinted polymer was prepared by using veterinary drug Sarafloxacin as template and itaconic acid (ITA) as functional monomer and ethylene glycol dimethacrylate (EDMA) as cross-linking agent. In order to increase the selectivity of imprinted polymers, cobalt ions are used to form metal-template complexes that can generate specific binding sites similar to metal-chelating antibodies. In protic solvents, the effect of different anions and cations on the adsorption performance of Sarafloxacin was studied. Because of the specific geometry of the complexes formed by Co2+ and sarafloxacin, the specific selectivity of the polymer to the template molecule was increased. It was found that the anion also participated in the recognition process and increased the specificity of the polymer to sarafloxacin. Scatchard analysis found that there are two binding sites, the maximum adsorption of 51.3 mmol g-1 and 32.6 mmol g-1. Compared with the conventionally synthesized molecularly imprinted polymers, this study achieved the synthesis and recognition of aqueous media. The results of selective adsorption experiments showed that the polymer has a higher selective binding ability to safranine.
Quinolones are a class of very important broad-spectrum antibiotics that have been rapidly developed in the past 20 years and are used extensively to treat, prevent and promote growth [1]. Its remnants in animal foods have become increasingly prominent. People’s long-term consumption of animal foods containing drug residues poses a potential threat to human health. In order to control the residues of quinolones in animal foods, it is necessary to develop a rapid, sensitive, and highly selective detection method for the determination of quinolones. One of the most important research directions is the development of good performance, high selectivity of the separation media for sample preparation and chromatography stationary phase, and the synthesis of this material is the main method of molecular imprinting technology.
(Molecularly Imprinted Polymer, MIP”>Molecular Imprinting Technology (MIT) simulates molecular recognition in nature, such as enzymes and substrates, antibodies and antigens, and uses target molecules as templates to synthesize molecularly imprinted polymers (MIPs) with specific molecular recognition functions. A technique [2], because MIP has the characteristics of predetermination, identification and practicality, and separation of enantiomers and isomers in many fields such as chromatography [3], solid phase extraction [4], chemistry Biomimetic sensors [5], simulated enzyme catalysis [6], clinical drug analysis [7] and membrane separation technology [8] have shown promising applications.
At present, the preparation and application of most molecularly imprinted polymers are limited to organic solvents, and the actual application environment for natural identification systems and MIPs is mostly aqueous systems, with the focus of research on MIP transferring from lipophilic organic small molecules. To hydrophilic compounds, the synthesis of molecular imprints in the aqueous phase is increasingly attracting people’s attention.
The recognition sites generated by molecularly imprinted polymers (MIPs) [9] during molecular imprinting often selectively bind imprinted molecules through forces such as ions, hydrogen bonds, and metal coordination. At present, there are many researches on hydrogen bonding and ion interaction in molecular imprinting, but less research on metal coordination. Among them, hydrogen bonding is the most commonly used interaction with molecular imprinting. However, the hydrogen bonds of the template interacting with the functional monomer are easily destroyed in the aqueous phase because the aqueous solvent competes with the template molecule for the functional monomer and thus is based on hydrogen. The effect of the molecularly imprinted polymer loses its high recognition ability in the highly polar aqueous or organic phase. When studying the interaction force in the process of biometric identification, it was found that the binding of biological or drug molecules and metal ions has a high degree of Specificity and mild binding and fracture conditions, which have an important role in the biometric system [10], such as heme, chlorophyll, vitamin B12 are complexes of iron, magnesium, and cobalt, respectively. Many drugs play a physiological role in the human body in the form of metal complexes, such as [Ca(EDTA)]2- can be used for the treatment of occupational lead poisoning, and cis-dichlorodiamine platinum(II) has good anticancer activity. 2,2-bipyridine has to play a specific pharmacological role through metal ions [11]. Therefore, studying the role of metal coordination in molecular recognition helps deepen the understanding of molecular imprinting and biometric processes and mechanisms. Because the metal coordination bond has its own unique advantages – strong acting, can be applied in strong polar solvents, so the molecular recognition characteristics of metal ion coordination MIP gradually attracted people’s attention. Fujii et al. [12] produced a metal-coordination molecularly imprinted polymer for the first time. The Co2+ ion binds amino acids to Schiff base ligands. The optical resolution of N-benzyl-D,L-proline is very high. The selectivity. Therefore, the study of selective recognition of metal coordination MIP has attracted people’s attention. Therefore, we synthesize metal-coordinated MIPs in the aqueous phase using the complexes formed by sarafloxacin and metal element (Co) as templates, and we focused on the effects of anions and cations on MIP affinity and binding.
1 Experimental section
1.1 Instruments and reagents
Instrument: analytical balance, magnetic stirrer(Shandong Hua Lu Electric instrument Co., Ltd.), constant temperature tank, a vacuum pump(Baoding high-tech Zone sunshine science instrument factory), sieve-like, constant temperature oscillator(Taicang science and education Equipment Factory), centrifuge(Beijing medical centrifuge factory), micro-hole filter, T6 UV spectrophotometer(Beijing Cape analysis universal Co., Ltd.), ultrasonic cleaner(Baoding ultrasonic cleaner factory).
Reagents : Sarafloxacin ( Taizhou Waigaoqiao Unicom Pharmaceutical Co., Ltd. ) , cobalt acetate ( Beijing Chemical Reagent Factory ) , ethylene glycol dimethacrylate (EDMA , Sigma-Aldrich Chemie) , itaconic acid ( Beijing Chemicals) Reagent company ) , acrylamide (MA , Tianjin Yongda Chemical Reagent Development Center ) , methacrylic acid (MAA , Sigma-Aldrich Chemie) , azobisisobutyronitrile (AIBN , Beijing Chemical Reagent Company ) , methanol ( Tianjin Regent Chemicals Co., Ltd. ) , Disodium EDTA (EDTA , Chemical Industry Reagent Branch of Tianjin Vertical and Horizontal Industry & Trade Co., Ltd. ) , Glacial acetic acid ( Beijing Beihua Fine Chemicals Co. , Ltd. ) , Ethanol ( Tianjin Meimei) Lin Industry and Trade Co., Ltd. ). The reagents used in the experiment were all analytically pure. The test water used was deionized water or high purity water.
1.2 molecularly imprinted polymer ( the MIP ) and non-imprinted polymers ( the NIP prepared) is
weighed Sarafloxacin (84.368mg , 0.2 mmol) , cobalt acetate (49.818 mg , 0.2 mmol) was dissolved in 6 ml of methanol / water (V / In V , 5:1) , sonicate solubilized for 2 h to fully complex and add 0.8 mmol of functional monomer ( itaconic acid, acrylamide, methacrylic acid, Table 1) , 1 ml of EDMA , and 6.6 mg of Dissolve the AIBN into a small test tube and incubate with argon for 15 min at 25 °C . Seal under vacuum and place in a 60 °C water bath for 24 h. Completion of the polymerization reaction, to obtain a rod-like loose polymer, into the 60 deg.] C vacuum oven dried, ground with a mortar, sieved to obtain 33.5-75 m m of the particulate polymer (the MIP) . Distillation extraction with 4:1 ethanol / acetic acid was used to elute sarafloxacin until no more safranine was detected at l =282.4 nm using a T6 UV spectrophotometer . Then, it was eluted with 0.1 mol L -1 EDTA solution for 2 h to remove Co(II) remaining in the polymer . Finally, it was eluted with methanol and transferred to a 60 °C vacuum oven for drying.
Acrylic acid and acrylamide polymers were prepared. Except for the replacement of functional monomers, the remaining steps were the same as MIP . The non-imprinted polymer ( NIP ) was prepared except that the metal coordination template ( cobalt (II) acetate and SAR) was not added . The rest of the procedure was the same as that of the MIP .
1.3 Balanced Binding Experiment of Polymer to Sarafloxacin 50 mg of imprinted polymer and comparative polymer were accurately weighed into a 25 ml Erlenmeyer flask, and 5 ml of sarafloxacin and cobalt acetate in methanol – water solution were added. (2 m mol ml -1 ) , put it in a constant temperature water bath shaker, and oscillate at 25 °C for 10 h , then take the sample of the adsorption solution in the centrifuge tube, take the centrifugation solution 0.1 ml after centrifugation , and dilute with water to 10.0 ml . The spectrophotometer was used for photometric analysis at 278.0 nm to determine the equilibrium concentration in the adsorption solution. The binding amount of the polymer was calculated based on the concentration of SAR in the solution before and after the combination. According to formula (1) , the imprinting factor IF of different functional monomer polymers is obtained.
IF = K MIP / K NIP = (Q MIP / C freeMIP) / (Q NIP / C freeNIP ) where K is the partition coefficient of different polymers, C free is the equilibrium concentration of adsorption, and Q is the adsorption amount of different polymers.
2 Results and Discussion
2.1 Synthesis of Co(II) coordination imprinted polymers
2.1.1 Selection of functional monomers
Table 1 Select monomer the Table 1 Selection of The Functional Monomers
|
polymer |
Template molecule |
Functional monomer |
Adsorption capacity Q(SAR) |
Imprinting factor IF |
|
MIP |
SAR+Co |
ITA |
24.2 |
2.87 |
|
NIP |
ITA |
9.15 |
||
|
MIP1 |
SAR+Co |
MAA |
54.53 |
1.54 |
|
NIP1 |
MAA |
39.05 |
||
|
MIP2 |
SAR+Co |
AM |
18.42 |
1.36 |
|
NIP2 |
AM |
15.55 |
Synthesis of molecularly imprinted polymers of SAR-Co complexes using methacrylic acid, itaconic acid, acrylamide as a functional monomer and ethylene glycol dimethacrylate as a cross-linking agent, static through ultraviolet spectrophotometer As shown in Table 1 , among the three functional monomers of the same amount, the polymer with itaconic acid as the functional monomer has the largest imprinting factor, so we chose itaconic acid as the functional monomer.
2.1.2 UV characteristics and synthesis mechanism of the prepolymerized solution

Figure 1 The absorbance of the aqueous solution of Sarafloxacin with itaconic acid
Fig.1 The curve of titrating ITA to Co(II)-SAR under the monitoring of UV spectrum
[Co( II)] = [SAR] = 0.02 [mu] mol · ml -1 , [the ITA] = 0, 0.010, 0.015, 0.020, 0.025 m mol · ml -1
The study of polymer recognition mechanism is very important for understanding the phenomenon of imprinting and recognition. Because the principle of molecular imprinting technology is based on the host-guest structure formed in the prepolymerized solution, the interaction between the template molecule and the functional monomer in the prepolymerized solution is performed. Characterization is very important. Therefore, a series of aqueous solutions of methanol with different concentration of Co(II)-SAR and different concentrations of itaconic acid were prepared. The UV absorbance changes of these solutions are shown in Fig. 1. There is a red shift near the absorption band at 286 nm . This is due to the decrease of electron cloud density due to the formation of the ITA-Co(II)-SAR complex. When [Co(II)-SAR]:[ITA]=1:1 , the maximum absorption wavelength of the solution no longer changes. From this can be derived synthesis of molecularly imprinted polymers schematic diagram 2.

Fig. 2 Molecular imprinting mechanism of itaconic acidFig. 2 Schematic representation of imprint formation
MIP 2.2 Factors affecting the adsorption characteristics of of Co 2+
2.2.1 The impact of Co 2+ on the ability to identify the ion concentration in order to examine MIP recognition capabilities, two comparative polymers was measured and the MIP of the SAR balanced by the amount of binding of cobalt acetate (II) concentration Effect of curve, FIG. 3 can be seen, MIP equilibrium binding was significantly greater than the amount of the NIP , showed that cobalt acetate (II) can be significantly increased in the presence of MIP selective recognition ability, when cobalt acetate concentration of 2 [mu] mol ml -1 when MIP has the strongest ability to identify. Then the equilibrium binding of MIP decreases with increasing cobalt acetate concentration. The reason is that MIP the Co (II) binding site has been Co (II) saturated solution of Co (II) of the SAR compete binding results MIP reduce binding amount.
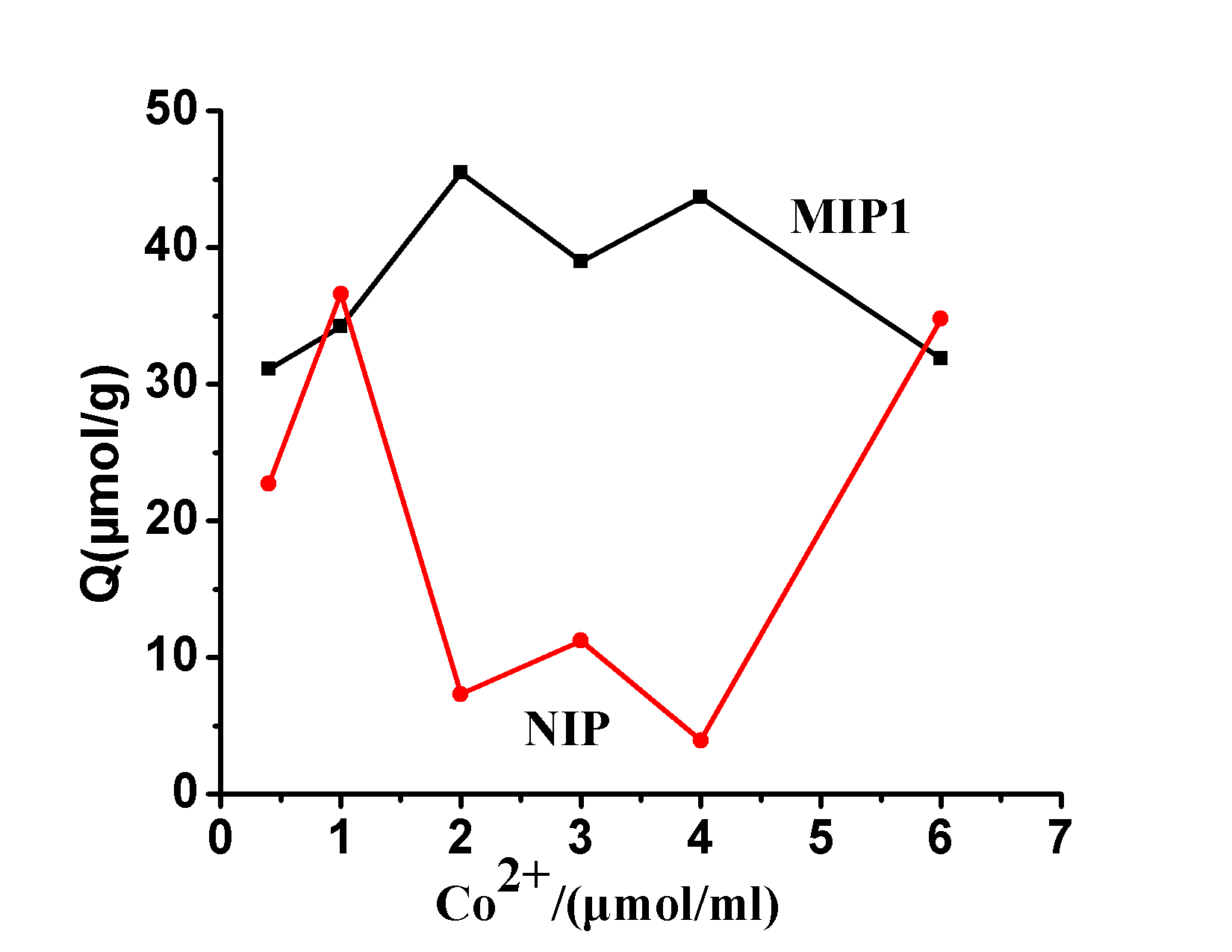
Figure 3. Co 2+ concentration on the amount of influence of polymer bound
Fig.3 Effect of Co 2+ Concentration on Polymers Binding Capacity
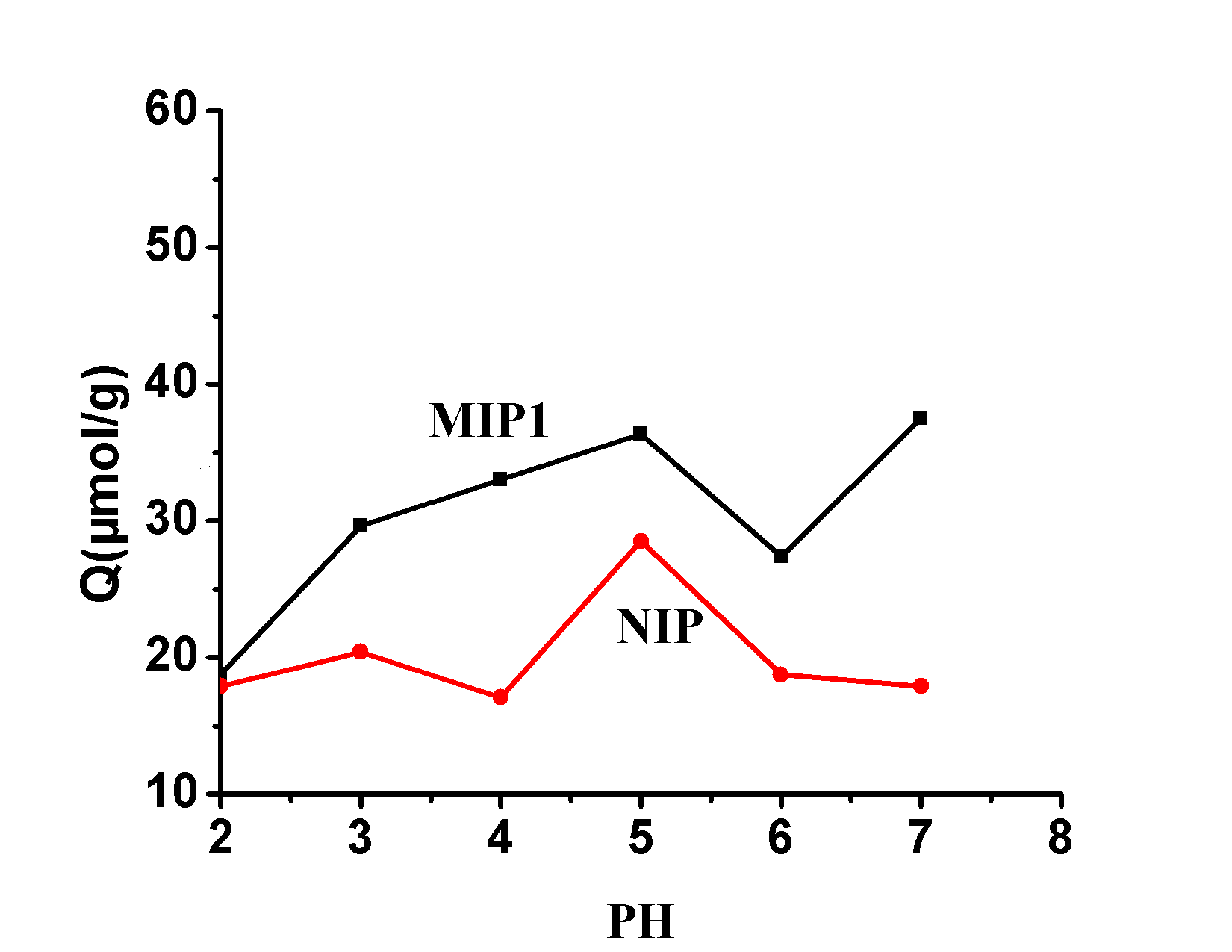
FIG.4 the pH value of its impact Imprinted
Fig.4 Effect of the pH ON Binding Capacity of Polymers
2.2.2 Effect of pH on MIP Recognition Ability
Since the molecularly imprinted polymer interacts with the target analyte via a metal bond, the effect of the pH of the solution on the amount of polymer bound was examined . As shown in FIG 4 , because when the [Co (II)] = 2 [mu] mol ml -1 , the pH =. 8 when Co (II) began to hydrolysis, so we pH 2-7 = within the range investigated. As can be seen from the figure, the binding of MIP and NIP to Sarafloxacin has an increasing trend with increasing pH . This is because at a low pH , the binding site is protonated and Co 2+ and binding sites are reduced. The force of the point, with the increase of pH value, is favorable for the formation of the Co 2+ -SAR complex, and it easily interacts with the binding site, and when the pH value is 7 , the maximum binding amount is reached. The binding of NIP to sarafloxacin due to nonselective specific adsorption and pHThere are ups and downs in the value. If you are following all the news about chemistry, you shouldn’t miss this post.
2.2.3 Effects of Cation on MIP Binding Amounts
Zinc acetate, copper acetate instead of cobalt acetate, these ions have similar ionic radii, r Zn ( II) = 0.074 nm , r Co ( II) = 0.072 nm , r Cu (II) ) = 0.071 nm , Table 2 shows thatthe selectivityof MIP incobalt acetate () medium isobviously greater than that of the other two media. The radius of these ions is similar.Compared with the other two ions, Co(II) is more likely toform a tetrahedral complex withitaconic acid and SAR , so that Co(II) can significantly increasethe selective recognition ability of MIP . This indicates that the selective binding of MIP is not the metal ion radius in the binding site but the geometry of the complex formed.
Table 2 Effects of Cation on MIP Binding Amount
Table 2 Effect of cations on binding capacity of polymers
| cation | Q( m mol·g-1) |
| Co(II) | 24.36 |
| Cu(II) | 20.85 |
| Zn(II) | 18.20 |
[SAR] = 2 m mol · ml -1 , [of Co (II)] = [a Cu (II)] = [a Zn (II)] = 2 m mol · ml -1
2.2.4 Anion MIP affect binding amount
Selected cobalt chloride (II) and cobalt sulfate (II) instead of cobalt acetate (II) were investigated on anion MIP affect ligand recognition ability. When the concentrations of Cl – , SO 4 -2, and Ac – are 2 μmol ml -1 , respectively, the binding of MIP to SAR is shown in Table 3. These indicate that anions are also involved in the binding process and form complexes, resulting in structure and properties. The difference, Ac – enables the MIP to generate effective selective recognition capabilities. And CI – on MIP affect binding without significant amounts of SO . 4 -2 large influence thereof, may be brought because the two different charges, resulting in different anions and cations resulting from the extent of binding.
Diagram 5 MIP ‘s Scatchard Curve
Fig.5 Scatchard plots of MIP
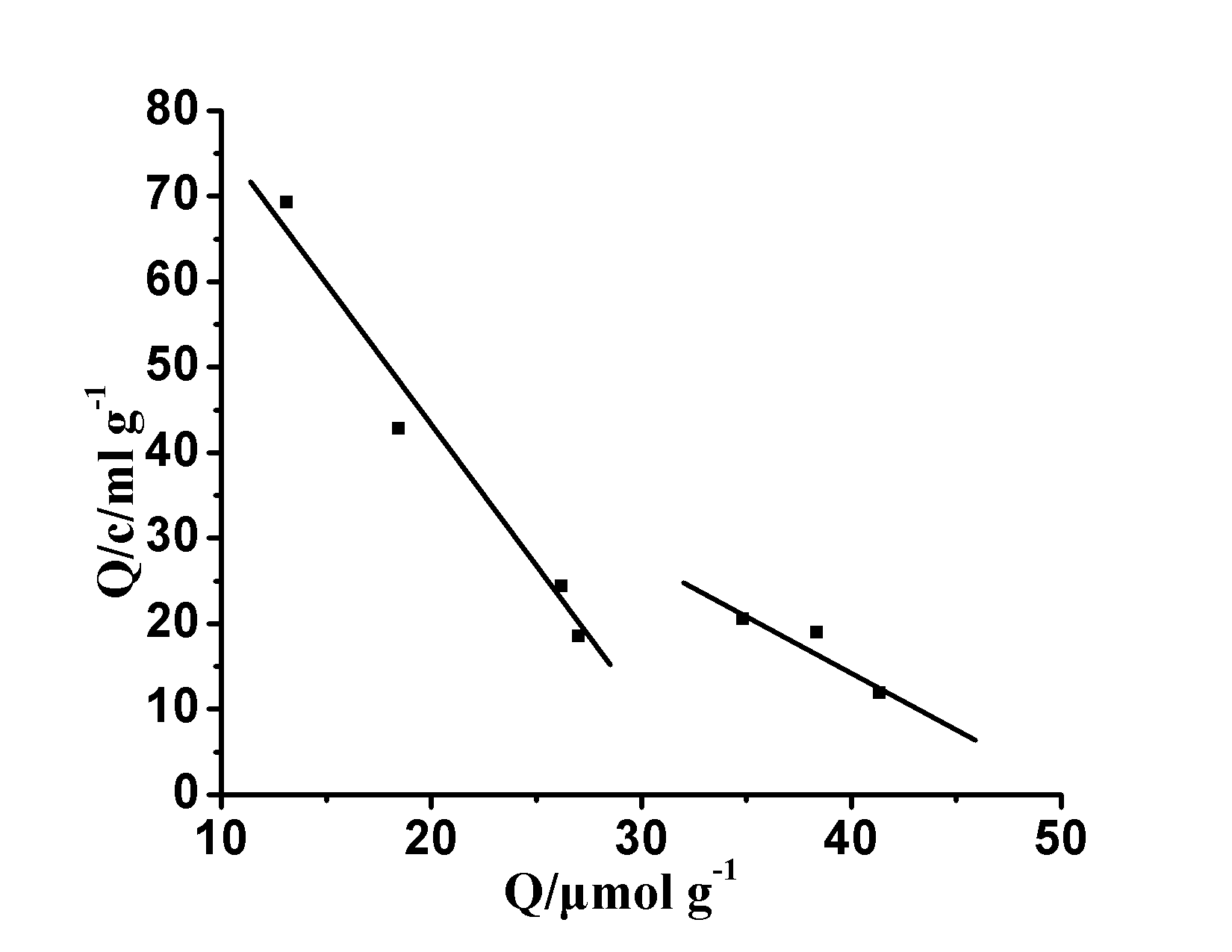
Table sub . 3 pair of female from MIP affect binding amount of
the Table. 3 Effect of Anions ON Binding Capacity of Polymers
| Anion | Ac – | Cl – | SO 4 -2 |
| Q( μmol·g-1) | 25.33 | 23.58 | 18.94 |
[SAR]=2 m mol·ml -1 , [Ac – ]=[Cl – ]=[SO 4 -2 ]= 2 m mol·ml -1
2.3 MIP of Scatchard analysis
in the molecule blot , conventional Scatchard binding model to evaluate the characteristics of the molecular imprinted polymer , of formula (2) is a Scatchard equation : Q / C = (Q max -Q is) / K D (2) where: K d equilibrium binding site on behalf dissociation constant, Q is the amount of binding, Q max apparent binding site on behalf of the maximum binding amount, C represents the equilibrium concentration of substrate, at Q / c on Q plot , can be obtained binding The combination of points K d and Q max . Ascan be seenfrom Figure 5 , Q/c is not linearto Q , indicating MIPThe binding effect on [Co(II)-SAR] 2+ is not uniform and has obvious multi-resistance. However, there is a clear linear relationship at both ends of the plot, which proves that there are two different types of binding sites for MIP within the concentration range of sarafloxacin studied . By fitting of the straight line portion in the figure , can be determined from the slope and intercept of the linear equation , a polymer of high affinity binding sites.
K D1 = 0.29 mmol ml -1 , Q MAX1 = 32.61 m mol G -1 , K d2 = 0.76 mmol ml -1
for the low affinity binding site and
Q max2 = 51.34 μ mol g -1.
2.4 imprinted polymer selective substrate
Selected sarafloxacin, ciprofloxacin as a substrate for selective adsorption experiments, the method was determined by equilibrium binding MIP binding amount studied MIP binding selectivity, the measurement results are shown in Table 4 . Ascan be seenfrom Table 4 ,the adsorption capacity of MIP for SAR is significantly greater than that of CIP , indicating that the polymerhas a good ability to identify SAR . From the molecular structure analysis, CIP isa fluorophenylsubstituted with cyclopropylon N1 by SAR , this substituent is smaller, butthe 3D dominant conformation of CIP moleculeshows that its cyclopropyl and main ring plane is vertical. and SAR -fluorophenyl is nearly parallel to the main plane of the ring, so CIP hindered too large to enter the PM binding site, thereby reducing the adsorption of the polymer to it.
Table 4 Polymer vs. Sarafloxacin and Its Structural AnalogsThe selective adsorption of
Table 4 adsorption of SAR and its analogues by MIP
| Substrate | Balanced concentration / mmol ml -1 | Q/ m mol g -1 | K d / ml g -1 | k |
| SAR | 1.76 | 24.20 | 13.75 | 1.52 |
| CIP | 1.83 | 16.51 | 9.02 |
K d , distribution coefficient, K d =Q/C free , where Q denotes adsorption capacity, C free denotes equilibrium concentration k , selectivity coefficient,
k = K d (SAR)/K d (CIP)
where K d (SAR) Denotes the partition coefficient of sarafloxacin, and K d (CIP) denotes the partition coefficient of ciprofloxacin.
3 Conclusions
The molecularly imprinted polymer of sarafloxacin was prepared in the aqueous phase by the coordination of cobalt ions and sarafloxacin. After experimental studies, MIP has a good adsorption effect on sarafloxacin. The selective recognition of the ligand by the metal coordination template polymer can be regulated by metal ions. This feature can meet the requirements of biosensors and man-made enzyme systems to control the recognition ability. Because the recognition force is the metal coordination force, metal coordination template polymers can be used in those systems where hydrogen bonds are easily destroyed or interfered with in water. Molecular recognition in the phase.
ACKNOWLEDGMENT
This work was supported by National Science Foundation of China (No. 20675024), Natural Science Foundation of Hebei Educational Committee (No. 2006407)
REFERENCES
[1] Li Junsuo , Qiu Yueming , Wang Chao . Analysis of veterinary drug residues . Shanghai : Shanghai Science and Technology Press , 2002, 257-258.
[2] Andersson LI J. Chromatogra. B, 2000, 745: 3-13.
[3 ] Peter S., Leif S., Lars IA, et al. Chromatographia, 2009, 69: 277-285.
[4] Jun M., Makoto O., Makoto T., et al. Anal. Commun., 1997, 34: 85-87
[5] Yonghong , high Zhixian , Chao Fuhuan . Analytical Chemistry , 2004, 32: 1407-1412.
[. 6] Polbornk, Severink Chem Commun, 1999, 24:… 2481-2491.
[. 7] Masanori N., Masaki O., Toshiaki N., et al. J. Pharmaceut. Biomed. Anal., 2005, 37: 231-237.
[8] Wang Guosong , Yang Xueqiong , side Yunshan , and the like . Chemical Research and Application , 2009, 21 is: 141-145.a
… [. 9] the ZF Xu, Xu L., Kuang the DZ, et Mater Sci Eng is Al, 2008. , 28: 1516-1521.
[10] Spirotg. Metal ions in biology. New York: Wiley Interscience, 1983, 76-77.
[11] Horky LL, Pluta RM, Book RJ J. Neurosurg, 1998, 88: 298- 303.
[12] Fujii Y., Kikuchi K., Matsutani K. Chem. Lett., 1984, 113: 1487-1490.
